 |
pyclustering
0.10.1
pyclustring is a Python, C++ data mining library.
|
 |
pyclustering
0.10.1
pyclustring is a Python, C++ data mining library.
|
PyClustering is an open source data mining library written in Python and C++ that provides a wide range of clustering algorithms and methods, including bio-inspired oscillatory networks. PyClustering is mostly focused on cluster analysis to make it more accessible and understandable for users. The library is distributed under the 3-Clause BSD License and provides a comprehensive interface that makes it easy to use in every project.
By default, the C++ part of the library is used for processing in order to achieve maximum performance. This is especially relevant for algorithms that are based on oscillatory networks, whose dynamics are governed by a system of differential equations. If support for a C++ compiler is not detected, PyClustering falls back to pure Python implementations of all kernels.
PyClustering consists of five general modules.
Cluster analysis algorithms and methods (module pyclustering.cluster):
Oscillatory and neural network models (module pyclustering.nnet):
Graph coloring algorithms (module pyclustering.gcolor):
Containers (module pyclustering.container):
Utils (pyclustering.utils) that can be used for analysis, visualization, etc.
The simplest way to install pyclustering library is to use pip:
The library can be built and installed manually. pyclustering's python code delegates computations to pyclustering C++ code that is represented by C++ pyclustering library: pyclustering.dll in case of Windows and libpyclustering.so in case of Linux and MacOS. There are three general ways to build C++ pyclustering:
ccore folder. Navigate to that folder: pyclustering/ccore.ccore.sln.Release configuration and corresponding platform (x64 or x86).pyclustering-shared project.
If you are using pyclustering library in a scientific paper, please, cite the library:
Novikov, A., 2019. PyClustering: Data Mining Library. Journal of Open Source Software, 4(36), p.1230. Available at: http://dx.doi.org/10.21105/joss.01230.
BibTeX entry:
This section contains few examples in order to demonstrate the interface of the library. The documentation contains examples for every algorithm/method/model/etc. More examples of a functionality can be found on a corresponding page of the function in this documentation.
The library provides intuitive and friendly interface. Here is an example how to perform cluster analysis using BIRCH algorithm:
Here is an how to perform cluster analysis using well-known K-Means algorithm:
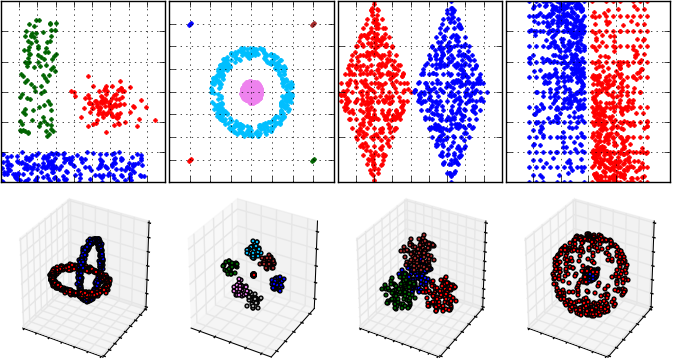
An example cluster analysis (that is performed by DBSCAN algorithm) for FCPS samples and visualization of results:
An example of Hodgkin-Huxley oscillatory network simulation with 6 oscillators. The first two oscillators have the same stimulus, as well as the third and fourth oscillators and the last two. Thus three synchronous ensembles are expected after simulation.