 |
pyclustering
0.10.1
pyclustring is a Python, C++ data mining library.
|
 |
pyclustering
0.10.1
pyclustring is a Python, C++ data mining library.
|
Class represents clustering algorithm K-Medoids (PAM algorithm). More...
Public Member Functions | |
| def | __init__ (self, data, initial_index_medoids, tolerance=0.0001, ccore=True, **kwargs) |
| Constructor of clustering algorithm K-Medoids. More... | |
| def | process (self) |
| Performs cluster analysis in line with rules of K-Medoids algorithm. More... | |
| def | predict (self, points) |
| Calculates the closest cluster to each point. More... | |
| def | get_clusters (self) |
| Returns list of allocated clusters, each cluster contains indexes of objects in list of data. More... | |
| def | get_medoids (self) |
| Returns list of medoids of allocated clusters represented by indexes from the input data. More... | |
| def | get_cluster_encoding (self) |
| Returns clustering result representation type that indicate how clusters are encoded. More... | |
Class represents clustering algorithm K-Medoids (PAM algorithm).
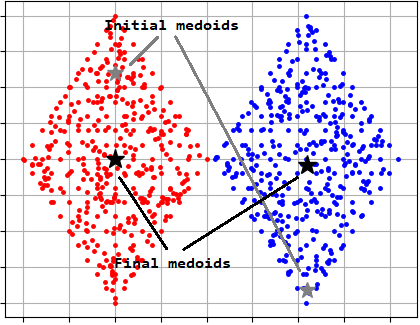
PAM is a partitioning clustering algorithm that uses the medoids instead of centers like in case of K-Means algorithm. Medoid is an object with the smallest dissimilarity to all others in the cluster. PAM algorithm complexity is \(O\left ( k\left ( n-k \right )^{2} \right )\).
There is an example where PAM algorithm is used to cluster 'TwoDiamonds' data:
Metric for calculation distance between points can be specified by parameter additional 'metric':
Distance matrix can be used instead of sequence of points to increase performance and for that purpose parameter 'data_type' should be used:
Definition at line 26 of file kmedoids.py.
| def pyclustering.cluster.kmedoids.kmedoids.__init__ | ( | self, | |
| data, | |||
| initial_index_medoids, | |||
tolerance = 0.0001, |
|||
ccore = True, |
|||
| ** | kwargs | ||
| ) |
Constructor of clustering algorithm K-Medoids.
| [in] | data | (list): Input data that is presented as list of points (objects), each point should be represented by list or tuple. |
| [in] | initial_index_medoids | (list): Indexes of intial medoids (indexes of points in input data). |
| [in] | tolerance | (double): Stop condition: if maximum value of distance change of medoids of clusters is less than tolerance than algorithm will stop processing. |
| [in] | ccore | (bool): If specified than CCORE library (C++ pyclustering library) is used for clustering instead of Python code. |
| [in] | **kwargs | Arbitrary keyword arguments (available arguments: 'metric', 'data_type', 'itermax'). |
Keyword Args:
Definition at line 100 of file kmedoids.py.
| def pyclustering.cluster.kmedoids.kmedoids.get_cluster_encoding | ( | self | ) |
Returns clustering result representation type that indicate how clusters are encoded.
Definition at line 248 of file kmedoids.py.
| def pyclustering.cluster.kmedoids.kmedoids.get_clusters | ( | self | ) |
Returns list of allocated clusters, each cluster contains indexes of objects in list of data.
Definition at line 224 of file kmedoids.py.
Referenced by pyclustering.samples.answer_reader.get_cluster_lengths(), and pyclustering.cluster.optics.optics.process().
| def pyclustering.cluster.kmedoids.kmedoids.get_medoids | ( | self | ) |
Returns list of medoids of allocated clusters represented by indexes from the input data.
Definition at line 236 of file kmedoids.py.
| def pyclustering.cluster.kmedoids.kmedoids.predict | ( | self, | |
| points | |||
| ) |
Calculates the closest cluster to each point.
| [in] | points | (array_like): Points for which closest clusters are calculated. |
An example how to calculate (or predict) the closest cluster to specified points.
Definition at line 178 of file kmedoids.py.
| def pyclustering.cluster.kmedoids.kmedoids.process | ( | self | ) |
Performs cluster analysis in line with rules of K-Medoids algorithm.
Definition at line 137 of file kmedoids.py.