 |
pyclustering
0.10.1
pyclustring is a Python, C++ data mining library.
|
 |
pyclustering
0.10.1
pyclustring is a Python, C++ data mining library.
|
Class represents clustering algorithm X-Means. More...
Public Member Functions | |
| def | __init__ (self, data, initial_centers=None, kmax=20, tolerance=0.001, criterion=splitting_type.BAYESIAN_INFORMATION_CRITERION, ccore=True, **kwargs) |
| Constructor of clustering algorithm X-Means. More... | |
| def | process (self) |
| Performs cluster analysis in line with rules of X-Means algorithm. More... | |
| def | predict (self, points) |
| Calculates the closest cluster to each point. More... | |
| def | get_clusters (self) |
| Returns list of allocated clusters, each cluster contains indexes of objects in list of data. More... | |
| def | get_centers (self) |
| Returns list of centers for allocated clusters. More... | |
| def | get_cluster_encoding (self) |
| Returns clustering result representation type that indicate how clusters are encoded. More... | |
| def | get_total_wce (self) |
| Returns sum of Euclidean Squared metric errors (SSE - Sum of Squared Errors). More... | |
Class represents clustering algorithm X-Means.
X-means clustering method starts with the assumption of having a minimum number of clusters, and then dynamically increases them. X-means uses specified splitting criterion to control the process of splitting clusters. Method K-Means++ can be used for calculation of initial centers.
CCORE implementation of the algorithm uses thread pool to parallelize the clustering process.
Here example how to perform cluster analysis using X-Means algorithm:
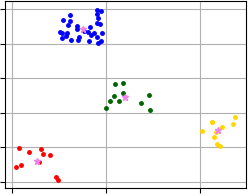
Visualization of clustering results that were obtained using code above and where X-Means algorithm allocates four clusters.
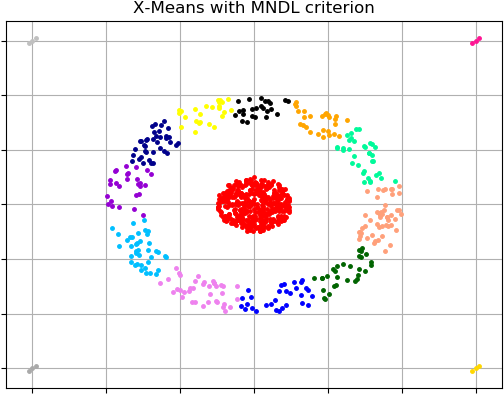
By default X-Means clustering algorithm uses Bayesian Information Criterion (BIC) to approximate the correct number of clusters. There is an example where another criterion Minimum Noiseless Description Length (MNDL) is used in order to find optimal amount of clusters:
As in many others algorithms, it is possible to specify metric that should be used for cluster analysis, for example, Chebyshev distance metric:
| def pyclustering.cluster.xmeans.xmeans.__init__ | ( | self, | |
| data, | |||
initial_centers = None, |
|||
kmax = 20, |
|||
tolerance = 0.001, |
|||
criterion = splitting_type.BAYESIAN_INFORMATION_CRITERION, |
|||
ccore = True, |
|||
| ** | kwargs | ||
| ) |
Constructor of clustering algorithm X-Means.
| [in] | data | (array_like): Input data that is presented as list of points (objects), each point should be represented by list or tuple. |
| [in] | initial_centers | (list): Initial coordinates of centers of clusters that are represented by list: [center1, center2, ...], if it is not specified then X-Means starts from the random center. |
| [in] | kmax | (uint): Maximum number of clusters that can be allocated. |
| [in] | tolerance | (double): Stop condition for each iteration: if maximum value of change of centers of clusters is less than tolerance than algorithm will stop processing. |
| [in] | criterion | (splitting_type): Type of splitting creation (by default splitting_type.BAYESIAN_INFORMATION_CRITERION). |
| [in] | ccore | (bool): Defines if C++ pyclustering library should be used instead of Python implementation. |
| [in] | **kwargs | Arbitrary keyword arguments (available arguments: repeat, random_state, metric, alpha, beta). |
Keyword Args:
1). With larger repeat values suggesting higher probability of finding global optimum.None, current system time is used).| def pyclustering.cluster.xmeans.xmeans.get_centers | ( | self | ) |
Returns list of centers for allocated clusters.
| def pyclustering.cluster.xmeans.xmeans.get_cluster_encoding | ( | self | ) |
Returns clustering result representation type that indicate how clusters are encoded.
| def pyclustering.cluster.xmeans.xmeans.get_clusters | ( | self | ) |
Returns list of allocated clusters, each cluster contains indexes of objects in list of data.
Definition at line 314 of file xmeans.py.
Referenced by pyclustering.samples.answer_reader.get_cluster_lengths().
| def pyclustering.cluster.xmeans.xmeans.get_total_wce | ( | self | ) |
Returns sum of Euclidean Squared metric errors (SSE - Sum of Squared Errors).
Sum of metric errors is calculated using distance between point and its center:
\[error=\sum_{i=0}^{N}euclidean_square_distance(x_{i}-center(x_{i}))\]
| def pyclustering.cluster.xmeans.xmeans.predict | ( | self, | |
| points | |||
| ) |
Calculates the closest cluster to each point.
| [in] | points | (array_like): Points for which closest clusters are calculated. |
An example how to calculate (or predict) the closest cluster to specified points.
| def pyclustering.cluster.xmeans.xmeans.process | ( | self | ) |
Performs cluster analysis in line with rules of X-Means algorithm.
Definition at line 206 of file xmeans.py.
Referenced by pyclustering.cluster.xmeans.xmeans.get_total_wce().