 |
pyclustering
0.10.1
pyclustring is a Python, C++ data mining library.
|
 |
pyclustering
0.10.1
pyclustring is a Python, C++ data mining library.
|
Class represents clustering algorithm OPTICS (Ordering Points To Identify Clustering Structure) with KD-tree optimization (ccore options is supported). More...
Public Member Functions | |
| def | __init__ (self, sample, eps, minpts, amount_clusters=None, ccore=True, **kwargs) |
| Constructor of clustering algorithm OPTICS. More... | |
| def | process (self) |
| Performs cluster analysis in line with rules of OPTICS algorithm. More... | |
| def | get_clusters (self) |
| Returns list of allocated clusters, where each cluster contains indexes of objects and each cluster is represented by list. More... | |
| def | get_noise (self) |
| Returns list of noise that contains indexes of objects that corresponds to input data. More... | |
| def | get_ordering (self) |
| Returns clustering ordering information about the input data set. More... | |
| def | get_optics_objects (self) |
| Returns OPTICS objects where each object contains information about index of point from processed data, core distance and reachability distance. More... | |
| def | get_radius (self) |
| Returns connectivity radius that is calculated and used for clustering by the algorithm. More... | |
| def | get_cluster_encoding (self) |
| Returns clustering result representation type that indicate how clusters are encoded. More... | |
Class represents clustering algorithm OPTICS (Ordering Points To Identify Clustering Structure) with KD-tree optimization (ccore options is supported).
OPTICS is a density-based algorithm. Purpose of the algorithm is to provide explicit clusters, but create clustering-ordering representation of the input data. Clustering-ordering information contains information about internal structures of data set in terms of density and proper connectivity radius can be obtained for allocation required amount of clusters using this diagram. In case of usage additional input parameter 'amount of clusters' connectivity radius should be bigger than real - because it will be calculated by the algorithms if requested amount of clusters is not allocated.
Clustering example using sample 'Chainlink':
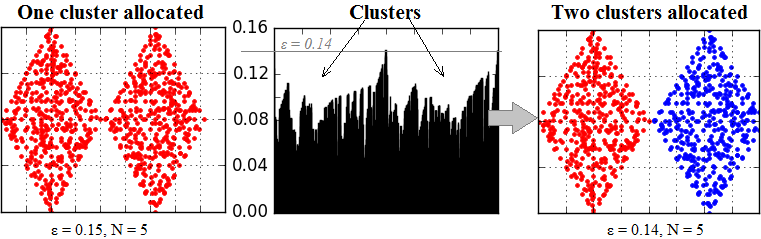
Amount of clusters that should be allocated can be also specified. In this case connectivity radius should be greater than real, for example:
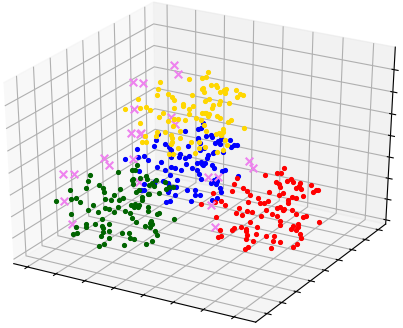
Here is an example where OPTICS extracts outliers from sample 'Tetra':
Visualization result of allocated clusters and outliers is presented on the image below:
| def pyclustering.cluster.optics.optics.__init__ | ( | self, | |
| sample, | |||
| eps, | |||
| minpts, | |||
amount_clusters = None, |
|||
ccore = True, |
|||
| ** | kwargs | ||
| ) |
Constructor of clustering algorithm OPTICS.
| [in] | sample | (list): Input data that is presented as a list of points (objects), where each point is represented by list or tuple. |
| [in] | eps | (double): Connectivity radius between points, points may be connected if distance between them less than the radius. |
| [in] | minpts | (uint): Minimum number of shared neighbors that is required for establishing links between points. |
| [in] | amount_clusters | (uint): Optional parameter where amount of clusters that should be allocated is specified. In case of usage 'amount_clusters' connectivity radius can be greater than real, in other words, there is place for mistake in connectivity radius usage. |
| [in] | ccore | (bool): if True than DLL CCORE (C++ solution) will be used for solving the problem. |
| [in] | **kwargs | Arbitrary keyword arguments (available arguments: 'data_type'). |
Keyword Args:
| def pyclustering.cluster.optics.optics.get_cluster_encoding | ( | self | ) |
Returns clustering result representation type that indicate how clusters are encoded.
| def pyclustering.cluster.optics.optics.get_clusters | ( | self | ) |
Returns list of allocated clusters, where each cluster contains indexes of objects and each cluster is represented by list.
Definition at line 508 of file optics.py.
Referenced by pyclustering.samples.answer_reader.get_cluster_lengths(), and pyclustering.cluster.optics.optics.process().
| def pyclustering.cluster.optics.optics.get_noise | ( | self | ) |
Returns list of noise that contains indexes of objects that corresponds to input data.
| def pyclustering.cluster.optics.optics.get_optics_objects | ( | self | ) |
Returns OPTICS objects where each object contains information about index of point from processed data, core distance and reachability distance.
| def pyclustering.cluster.optics.optics.get_ordering | ( | self | ) |
Returns clustering ordering information about the input data set.
Clustering ordering of data-set contains the information about the internal clustering structure in line with connectivity radius.
Definition at line 540 of file optics.py.
Referenced by pyclustering.cluster.optics.optics.process().
| def pyclustering.cluster.optics.optics.get_radius | ( | self | ) |
Returns connectivity radius that is calculated and used for clustering by the algorithm.
Connectivity radius may be changed only in case of usage additional parameter of the algorithm - amount of clusters for allocation.
| def pyclustering.cluster.optics.optics.process | ( | self | ) |
Performs cluster analysis in line with rules of OPTICS algorithm.