 |
 |
Oscillatory Neural Network with central element based on Hodgkin-Huxley neuron model. More...
 Inheritance diagram for pyclustering.nnet.hhn.hhn_network:
Inheritance diagram for pyclustering.nnet.hhn.hhn_network: Collaboration diagram for pyclustering.nnet.hhn.hhn_network:
Collaboration diagram for pyclustering.nnet.hhn.hhn_network:Public Member Functions | |
| def | __init__ (self, num_osc, stimulus=None, parameters=None, type_conn=None, type_conn_represent=conn_represent.MATRIX, ccore=True) |
| Constructor of oscillatory network based on Hodgkin-Huxley neuron model. More... | |
| def | __del__ (self) |
| Destroy dynamically allocated oscillatory network instance in case of CCORE usage. | |
| def | simulate (self, steps, time, solution=solve_type.RK4) |
| Performs static simulation of oscillatory network based on Hodgkin-Huxley neuron model. More... | |
| def | simulate_static (self, steps, time, solution=solve_type.RK4) |
| Performs static simulation of oscillatory network based on Hodgkin-Huxley neuron model. More... | |
| def | hnn_state (self, inputs, t, argv) |
| Returns new values of excitatory and inhibitory parts of oscillator and potential of oscillator. More... | |
| def | allocate_sync_ensembles (self, tolerance=0.1) |
| Allocates clusters in line with ensembles of synchronous oscillators where each. More... | |
 Public Member Functions inherited from pyclustering.nnet.network Public Member Functions inherited from pyclustering.nnet.network | |
| def | height (self) |
| Height of the network grid (that is defined by amout of oscillators in each column), this value is zero in case of non-grid structure. More... | |
| def | width (self) |
| Width of the network grid, this value is zero in case of non-grid structure. More... | |
| def | structure (self) |
| Type of network structure that is used for connecting oscillators. | |
| def | __init__ (self, num_osc, type_conn=conn_type.ALL_TO_ALL, conn_repr=conn_represent.MATRIX, height=None, width=None) |
| Constructor of the network. More... | |
| def | __len__ (self) |
| Returns size of the network that is defined by amount of oscillators. | |
| def | has_connection (self, i, j) |
| Returns True if there is connection between i and j oscillators and False - if connection doesn't exist. More... | |
| def | set_connection (self, i, j) |
| Couples two specified oscillators in the network with dynamic connections. More... | |
| def | get_neighbors (self, index) |
| Finds neighbors of the oscillator with specified index. More... | |
Oscillatory Neural Network with central element based on Hodgkin-Huxley neuron model.
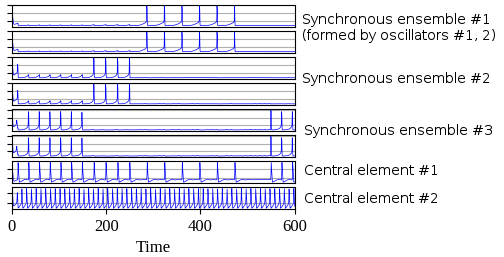
Interaction between oscillators is performed via central element (no connection between oscillators that are called as peripheral). Peripheral oscillators receive external stimulus. Central element consist of two oscillators: the first is used for synchronization some ensemble of oscillators and the second controls synchronization of the first central oscillator with various ensembles.
Usage example where oscillatory network with 6 oscillators is used for simulation. The first two oscillators have the same stimulus, as well as the third and fourth oscillators and the last two. Thus three synchronous ensembles are expected after simulation.
To increase performance CCORE can be used, for that purpose special flag should be specified when network is constructed:
There is visualized result of simulation where three synchronous ensembles of oscillators can be observed. The first and the second oscillators form the first ensemble, the third and the fourth form the second ensemble and the last two oscillators form the third ensemble.
| def pyclustering.nnet.hhn.hhn_network.__init__ | ( | self, | |
| num_osc, | |||
stimulus = None, |
|||
parameters = None, |
|||
type_conn = None, |
|||
type_conn_represent = conn_represent.MATRIX, |
|||
ccore = True |
|||
| ) |
Constructor of oscillatory network based on Hodgkin-Huxley neuron model.
| [in] | num_osc | (uint): Number of peripheral oscillators in the network. |
| [in] | stimulus | (list): List of stimulus for oscillators, number of stimulus should be equal to number of peripheral oscillators. |
| [in] | parameters | (hhn_parameters): Parameters of the network. |
| [in] | type_conn | (conn_type): Type of connections between oscillators in the network (ignored for this type of network). |
| [in] | type_conn_represent | (conn_represent): Internal representation of connection in the network: matrix or list. |
| [in] | ccore | (bool): If 'True' then CCORE is used (C/C++ implementation of the model). |
| def pyclustering.nnet.hhn.hhn_network.allocate_sync_ensembles | ( | self, | |
tolerance = 0.1 |
|||
| ) |
Allocates clusters in line with ensembles of synchronous oscillators where each.
Synchronous ensemble corresponds to only one cluster.
| [in] | tolerance | (double): maximum error for allocation of synchronous ensemble oscillators. |
Definition at line 555 of file hhn.py.
Referenced by pyclustering.gcolor.hysteresis.hysteresis_analyser.allocate_clusters(), pyclustering.cluster.syncnet.syncnet_analyser.allocate_clusters(), and pyclustering.gcolor.sync.syncgcolor_analyser.allocate_color_clusters().
| def pyclustering.nnet.hhn.hhn_network.hnn_state | ( | self, | |
| inputs, | |||
| t, | |||
| argv | |||
| ) |
Returns new values of excitatory and inhibitory parts of oscillator and potential of oscillator.
| [in] | inputs | (list): States of oscillator for integration [v, m, h, n] (see description below). |
| [in] | t | (double): Current time of simulation. |
| [in] | argv | (tuple): Extra arguments that are not used for integration - index of oscillator. |
Definition at line 468 of file hhn.py.
Referenced by pyclustering.nnet.hhn.hhn_network.simulate_static().
| def pyclustering.nnet.hhn.hhn_network.simulate | ( | self, | |
| steps, | |||
| time, | |||
solution = solve_type.RK4 |
|||
| ) |
Performs static simulation of oscillatory network based on Hodgkin-Huxley neuron model.
Output dynamic is sensible to amount of steps of simulation and solver of differential equation. Python implementation uses 'odeint' from 'scipy', CCORE uses classical RK4 and RFK45 methods, therefore in case of CCORE HHN (Hodgkin-Huxley network) amount of steps should be greater than in case of Python HHN.
| [in] | steps | (uint): Number steps of simulations during simulation. |
| [in] | time | (double): Time of simulation. |
| [in] | solution | (solve_type): Type of solver for differential equations. |
| def pyclustering.nnet.hhn.hhn_network.simulate_static | ( | self, | |
| steps, | |||
| time, | |||
solution = solve_type.RK4 |
|||
| ) |
Performs static simulation of oscillatory network based on Hodgkin-Huxley neuron model.
Output dynamic is sensible to amount of steps of simulation and solver of differential equation. Python implementation uses 'odeint' from 'scipy', CCORE uses classical RK4 and RFK45 methods, therefore in case of CCORE HHN (Hodgkin-Huxley network) amount of steps should be greater than in case of Python HHN.
| [in] | steps | (uint): Number steps of simulations during simulation. |
| [in] | time | (double): Time of simulation. |
| [in] | solution | (solve_type): Type of solver for differential equations. |
Definition at line 289 of file hhn.py.
Referenced by pyclustering.nnet.hhn.hhn_network.simulate(), pyclustering.nnet.hysteresis.hysteresis_network.simulate(), pyclustering.nnet.syncpr.syncpr.simulate(), and pyclustering.nnet.sync.sync_network.simulate().