 |
 |
PyClustering library is a collection of cluster analysis, graph coloring, travelling salesman problem algorithms, oscillatory and neural network models, containers, tools for visualization and result analysis, etc. High performance is ensured by CCORE library that is a part of the pyclustering library where almost the same algorithms, models, tools are implemented. There is ability to use python code implementation only or CCORE (C/C++) implementation using special flag. CCORE library does not use python.h interface to communicate with python code due to requirement to save ability to use CCORE library or C/C++ code part of CCORE in other projects.
PyClustering consists of six general modules where the algorithms, models, tools are placed:
Cluster analysis algorithms and methods (module pyclustering.cluster):
Oscillatory and neural network models (module pyclustering.nnet):
Graph coloring algorithms (module pyclustering.gcolor):
Containers (module pyclustering.container):
Utils that can be used for analysis, visualization, etc are placed in module pyclustering.utils.
The simplest way to install pyclustering library is to use pip:
The library can be compiled and manually installed on linux machine wherever you want:
The library CCORE for 64-bit windows is distributed with pyclustering library so there is no need to re-built it. If you want to re-built CCORE library you can open CCORE Microsoft Visual Studio project that is located in ccore/ folder and compile it.
The library provides intuitive and friendly interface, cluster analysis can be easily performed:
Clustering algorithms can be used for image processing:
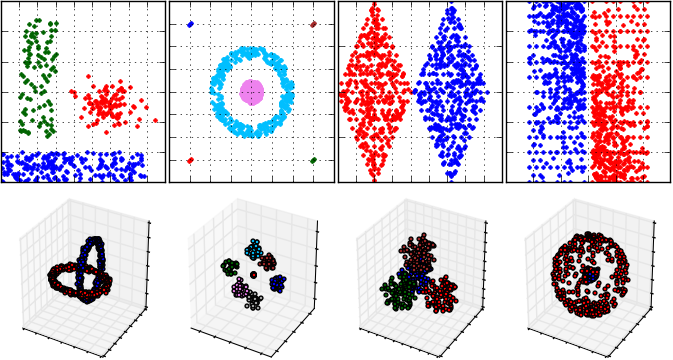
An example cluster analysis (that is performed by DBSCAN algorithm) for FCPS samples and visualization of results:
Simulation of oscillatory network based on Hodgkin-Huxley neuron model where six synchronous ensembles of oscillators are formed. It means that three features from input data are allocated where each feature is encoded by only one ensemble.