 |
 |
Expectation-Maximization clustering algorithm for Gaussian Mixture Model (GMM). More...
Public Member Functions | |
| def | __init__ (self, data, amount_clusters, means=None, variances=None, observer=None, tolerance=0.00001, iterations=100) |
| Initializes Expectation-Maximization algorithm for cluster analysis. More... | |
| def | process (self) |
| Run clustering process of the algorithm. More... | |
| def | get_clusters (self) |
| def | get_centers (self) |
| def | get_covariances (self) |
| def | get_probabilities (self) |
| Returns 2-dimensional list with belong probability of each object from data to cluster correspondingly, where that first index is for cluster and the second is for point. More... | |
Expectation-Maximization clustering algorithm for Gaussian Mixture Model (GMM).
The algorithm provides only clustering services (unsupervised learning). Here an example of data clustering process:
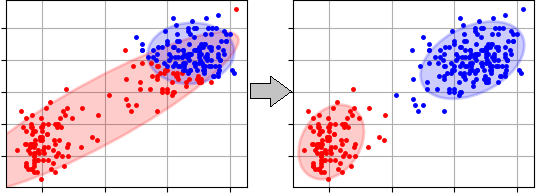
Here is clustering results of the Expectation-Maximization clustering algorithm where popular sample 'OldFaithful' was used. Initial random means and covariances were used in the example. The first step is presented on the left side of the figure and final result (the last step) is on the right side:
| def pyclustering.cluster.ema.ema.__init__ | ( | self, | |
| data, | |||
| amount_clusters, | |||
means = None, |
|||
variances = None, |
|||
observer = None, |
|||
tolerance = 0.00001, |
|||
iterations = 100 |
|||
| ) |
Initializes Expectation-Maximization algorithm for cluster analysis.
| [in] | data | (list): Dataset that should be analysed and where each point (object) is represented by the list of coordinates. |
| [in] | amount_clusters | (uint): Amount of clusters that should be allocated. |
| [in] | means | (list): Initial means of clusters (amount of means should be equal to amount of clusters for allocation). If this parameter is 'None' then K-Means algorithm with K-Means++ method will be used for initialization by default. |
| [in] | variances | (list): Initial cluster variances (or covariances in case of multi-dimensional data). Amount of covariances should be equal to amount of clusters that should be allocated. If this parameter is 'None' then K-Means algorithm with K-Means++ method will be used for initialization by default. |
| [in] | observer | (ema_observer): Observer for gathering information about clustering process. |
| [in] | tolerance | (float): Defines stop condition of the algorithm (when difference between current and previous log-likelihood estimation is less then 'tolerance' then clustering is over). |
| [in] | iterations | (uint): Additional stop condition parameter that defines maximum number of steps that can be performed by the algorithm during clustering process. |
| def pyclustering.cluster.ema.ema.get_centers | ( | self | ) |
| def pyclustering.cluster.ema.ema.get_clusters | ( | self | ) |
Definition at line 548 of file ema.py.
Referenced by pyclustering.samples.answer_reader.get_cluster_lengths(), and pyclustering.cluster.optics.optics.process().
| def pyclustering.cluster.ema.ema.get_covariances | ( | self | ) |
| def pyclustering.cluster.ema.ema.get_probabilities | ( | self | ) |
Returns 2-dimensional list with belong probability of each object from data to cluster correspondingly, where that first index is for cluster and the second is for point.
| def pyclustering.cluster.ema.ema.process | ( | self | ) |
Run clustering process of the algorithm.
This method should be called before call 'get_clusters()'.