 |
 |
Class implements CLIQUE grid based clustering algorithm. More...
Public Member Functions | |
| def | __init__ (self, data, amount_intervals, density_threshold, kwargs) |
| Create CLIQUE clustering algorithm. More... | |
| def | process (self) |
| Performs clustering process in line with rules of CLIQUE clustering algorithm. More... | |
| def | get_clusters (self) |
| Returns allocated clusters. More... | |
| def | get_noise (self) |
| Returns allocated noise. More... | |
| def | get_cells (self) |
| Returns CLIQUE blocks that are formed during clustering process. More... | |
| def | get_cluster_encoding (self) |
| Returns clustering result representation type that indicate how clusters are encoded. More... | |
Class implements CLIQUE grid based clustering algorithm.
CLIQUE automatically finds subspaces with high-density clusters. It produces identical results irrespective of the order in which the input records are presented and it does not presume any canonical distribution for input data [1].
Here is an example where data in two-dimensional space is clustered using CLIQUE algorithm:
In this example 6 clusters are allocated including four small cluster where each such small cluster consists of three points. There are visualized clustering results - grid that has been formed by CLIQUE algorithm with density and clusters itself:
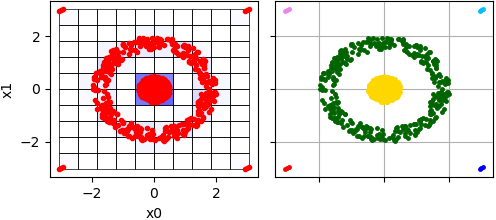
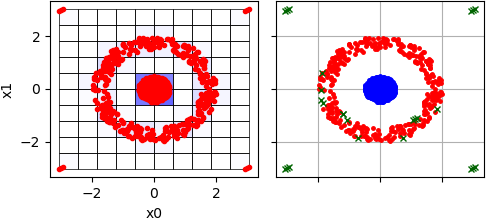
Sometimes such small clusters should be considered as outliers taking into account fact that two clusters in the central are relatively huge. To treat them as a noise threshold value should be increased:
Two clusters are allocated, but in this case some points in cluster-"circle" are also considered as outliers, because CLIQUE operates with blocks, not with points:
| def pyclustering.cluster.clique.clique.__init__ | ( | self, | |
| data, | |||
| amount_intervals, | |||
| density_threshold, | |||
| kwargs | |||
| ) |
Create CLIQUE clustering algorithm.
| [in] | data | (list): Input data (list of points) that should be clustered. |
| [in] | amount_intervals | (uint): Amount of intervals in each dimension that defines amount of CLIQUE blocks as
|
| [in] | density_threshold | (uint): Minimum number of points that should contain CLIQUE block to consider its points as non-outliers. |
| [in] | **kwargs | Arbitrary keyword arguments (available arguments: 'ccore'). |
Keyword Args:
| def pyclustering.cluster.clique.clique.get_cells | ( | self | ) |
Returns CLIQUE blocks that are formed during clustering process.
CLIQUE blocks can be used for visualization purposes. Each CLIQUE block contain its logical location in grid, spatial location in data space and points that belong to block.
| def pyclustering.cluster.clique.clique.get_cluster_encoding | ( | self | ) |
Returns clustering result representation type that indicate how clusters are encoded.
| def pyclustering.cluster.clique.clique.get_clusters | ( | self | ) |
Returns allocated clusters.
Definition at line 543 of file clique.py.
Referenced by pyclustering.samples.answer_reader.get_cluster_lengths(), and pyclustering.cluster.optics.optics.process().
| def pyclustering.cluster.clique.clique.get_noise | ( | self | ) |
Returns allocated noise.
| def pyclustering.cluster.clique.clique.process | ( | self | ) |
Performs clustering process in line with rules of CLIQUE clustering algorithm.