 |
 |
Class represents Genetic clustering algorithm. More...
Public Member Functions | |
| def | __init__ (self, data, count_clusters, chromosome_count, population_count, kwargs) |
| Initialize genetic clustering algorithm for cluster analysis. More... | |
| def | process (self) |
| Perform clustering procedure in line with rule of genetic clustering algorithm. More... | |
| def | get_observer (self) |
| Returns genetic algorithm observer. | |
| def | get_clusters (self) |
| Returns list of allocated clusters, each cluster contains indexes of objects from the data. More... | |
Class represents Genetic clustering algorithm.
The searching capability of genetic algorithms is exploited in order to search for appropriate cluster centres.
Example of clustering using genetic algorithm:
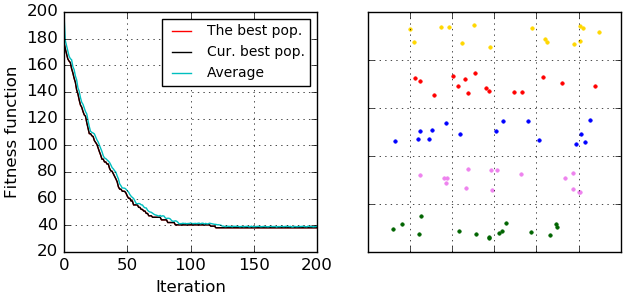
There is an example of clustering results (fitness function evolution and allocated clusters) that were visualized by 'ga_visualizer':
| def pyclustering.cluster.ga.genetic_algorithm.__init__ | ( | self, | |
| data, | |||
| count_clusters, | |||
| chromosome_count, | |||
| population_count, | |||
| kwargs | |||
| ) |
Initialize genetic clustering algorithm for cluster analysis.
| [in] | data | (numpy.array|list): Input data for clustering that is represented by two dimensional array where each row is a point, for example, [[0.0, 2.1], [0.1, 2.0], [-0.2, 2.4]]. |
| [in] | count_clusters | (uint): Amount of clusters that should be allocated in the data. |
| [in] | chromosome_count | (uint): Amount of chromosomes in each population. |
| [in] | population_count | (uint): Amount of populations. |
| [in] | **kwargs | Arbitrary keyword arguments (available arguments: 'count_mutation_gens', 'coeff_mutation_count', 'select_coeff', 'crossover_rate', 'observer', 'random_state'). |
Keyword Args:
None, current system time is used). | def pyclustering.cluster.ga.genetic_algorithm.get_clusters | ( | self | ) |
Returns list of allocated clusters, each cluster contains indexes of objects from the data.
Definition at line 495 of file ga.py.
Referenced by pyclustering.samples.answer_reader.get_cluster_lengths(), and pyclustering.cluster.optics.optics.process().
| def pyclustering.cluster.ga.genetic_algorithm.process | ( | self | ) |
Perform clustering procedure in line with rule of genetic clustering algorithm.