 |
 |
Represents Silhouette method that is used interpretation and validation of consistency. More...
Public Member Functions | |
| def | __init__ (self, data, clusters, kwargs) |
| Initializes Silhouette method for analysis. More... | |
| def | process (self) |
| Calculates Silhouette score for each object from input data. More... | |
| def | get_score (self) |
| Returns Silhouette score for each object from input data. More... | |
Represents Silhouette method that is used interpretation and validation of consistency.
The silhouette value is a measure of how similar an object is to its own cluster compared to other clusters. Be aware that silhouette method is applicable for K algorithm family, such as K-Means, K-Medians, K-Medoids, X-Means, etc., not not applicable for DBSCAN, OPTICS, CURE, etc. The Silhouette value is calculated using following formula:
![\[s\left ( i \right )=\frac{ b\left ( i \right ) - a\left ( i \right ) }{ max\left \{ a\left ( i \right ), b\left ( i \right ) \right \}}\]](../../form_11.png)
where 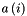 - is average distance from object i to objects in its own cluster,
- is average distance from object i to objects in its own cluster, 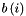 - is average distance from object i to objects in the nearest cluster (the appropriate among other clusters).
- is average distance from object i to objects in the nearest cluster (the appropriate among other clusters).
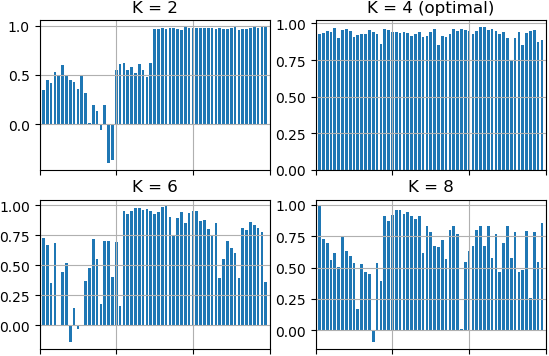
Here is an example where Silhouette score is calculated for K-Means's clustering result:
Let's perform clustering of the same sample by K-Means algorithm using different K values (2, 4, 6 and 8) and estimate clustering results using Silhouette method.
There is visualized results that were done by Silhouette method. K = 4 is the optimal amount of clusters in line with Silhouette method because the score for each point is close to 1.0 and the average score for K = 4 is biggest value among others K.
Definition at line 45 of file silhouette.py.
| def pyclustering.cluster.silhouette.silhouette.__init__ | ( | self, | |
| data, | |||
| clusters, | |||
| kwargs | |||
| ) |
Initializes Silhouette method for analysis.
| [in] | data | (array_like): Input data that was used for cluster analysis and that is presented as list of points or distance matrix (defined by parameter 'data_type', by default data is considered as a list of points). |
| [in] | clusters | (list): Clusters that have been obtained after cluster analysis. |
| [in] | **kwargs | Arbitrary keyword arguments (available arguments: 'metric'). |
Keyword Args:
Definition at line 144 of file silhouette.py.
| def pyclustering.cluster.silhouette.silhouette.get_score | ( | self | ) |
Returns Silhouette score for each object from input data.
Definition at line 217 of file silhouette.py.
| def pyclustering.cluster.silhouette.silhouette.process | ( | self | ) |
Calculates Silhouette score for each object from input data.
Definition at line 183 of file silhouette.py.