 |
pyclustering
0.10.1
pyclustring is a Python, C++ data mining library.
|
 |
pyclustering
0.10.1
pyclustring is a Python, C++ data mining library.
|
Represents self-organized feature map (SOM). More...
Public Member Functions | |
| def | size (self) |
| Return size of self-organized map that is defined by total number of neurons. More... | |
| def | weights (self) |
| Return weight of each neuron. More... | |
| def | awards (self) |
| Return amount of captured objects by each neuron after training. More... | |
| def | capture_objects (self) |
| Returns indexes of captured objects by each neuron. More... | |
| def | __init__ (self, rows, cols, conn_type=type_conn.grid_eight, parameters=None, ccore=True) |
| Constructor of self-organized map. More... | |
| def | __del__ (self) |
| Destructor of the self-organized feature map. | |
| def | __len__ (self) |
| Returns size of the network that defines by amount of neuron in it. More... | |
| def | __getstate__ (self) |
| def | __setstate__ (self, som_state) |
| def | train (self, data, epochs, autostop=False) |
| Trains self-organized feature map (SOM). More... | |
| def | simulate (self, input_pattern) |
| Processes input pattern (no learining) and returns index of neuron-winner. More... | |
| def | get_winner_number (self) |
| Calculates number of winner at the last step of learning process. More... | |
| def | show_distance_matrix (self) |
| Shows gray visualization of U-matrix (distance matrix). More... | |
| def | get_distance_matrix (self) |
| Calculates distance matrix (U-matrix). More... | |
| def | show_density_matrix (self, surface_divider=20.0) |
| Show density matrix (P-matrix) using kernel density estimation. More... | |
| def | get_density_matrix (self, surface_divider=20.0) |
| Calculates density matrix (P-Matrix). More... | |
| def | show_winner_matrix (self) |
| Show a winner matrix where each element corresponds to neuron and value represents amount of won objects from input data-space at the last training iteration. More... | |
| def | show_network (self, awards=False, belongs=False, coupling=True, dataset=True, marker_type='o') |
| Shows neurons in the dimension of data. More... | |
Represents self-organized feature map (SOM).
The self-organizing feature map (SOM) method is a powerful tool for the visualization of of high-dimensional data. It converts complex, nonlinear statistical relationships between high-dimensional data into simple geometric relationships on a low-dimensional display.
ccore option can be specified in order to control using C++ implementation of pyclustering library. By default C++ implementation is on. C++ implementation improves performance of the self-organized feature map.
Example:
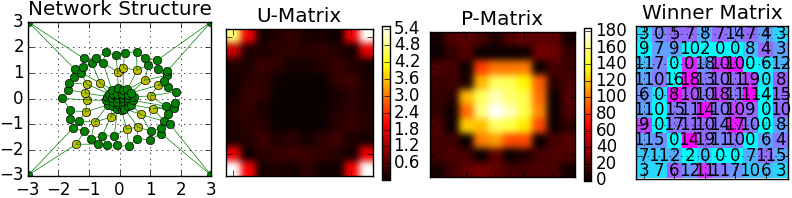
There is a visualization of 'Target' sample that was done by the self-organized feature map:
| def pyclustering.nnet.som.som.__init__ | ( | self, | |
| rows, | |||
| cols, | |||
conn_type = type_conn.grid_eight, |
|||
parameters = None, |
|||
ccore = True |
|||
| ) |
Constructor of self-organized map.
| [in] | rows | (uint): Number of neurons in the column (number of rows). |
| [in] | cols | (uint): Number of neurons in the row (number of columns). |
| [in] | conn_type | (type_conn): Type of connection between oscillators in the network (grid four, grid eight, honeycomb, function neighbour). |
| [in] | parameters | (som_parameters): Other specific parameters. |
| [in] | ccore | (bool): If True simulation is performed by CCORE library (C++ implementation of pyclustering). |
| def pyclustering.nnet.som.som.__getstate__ | ( | self | ) |
| def pyclustering.nnet.som.som.__len__ | ( | self | ) |
| def pyclustering.nnet.som.som.__setstate__ | ( | self, | |
| som_state | |||
| ) |
| def pyclustering.nnet.som.som.awards | ( | self | ) |
| def pyclustering.nnet.som.som.capture_objects | ( | self | ) |
Returns indexes of captured objects by each neuron.
For example, a network with size 2x2 has been trained on a sample with five objects. Suppose neuron #1 won an object with index 1, neuron #2 won objects 0, 3, 4, neuron #3 did not won anything and finally neuron #4 won an object with index 2. Thus, for this example we will have the following output [[1], [0, 3, 4], [], [2]].
| def pyclustering.nnet.som.som.get_density_matrix | ( | self, | |
surface_divider = 20.0 |
|||
| ) |
Calculates density matrix (P-Matrix).
| [in] | surface_divider | (double): Divider in each dimension that affect radius for density measurement. |
Definition at line 767 of file som.py.
Referenced by pyclustering.nnet.som.som.show_density_matrix().
| def pyclustering.nnet.som.som.get_distance_matrix | ( | self | ) |
Calculates distance matrix (U-matrix).
The U-Matrix visualizes based on the distance in input space between a weight vector and its neighbors on map.
Definition at line 717 of file som.py.
Referenced by pyclustering.nnet.som.som.show_distance_matrix().
| def pyclustering.nnet.som.som.get_winner_number | ( | self | ) |
| def pyclustering.nnet.som.som.show_density_matrix | ( | self, | |
surface_divider = 20.0 |
|||
| ) |
Show density matrix (P-matrix) using kernel density estimation.
| [in] | surface_divider | (double): Divider in each dimension that affect radius for density measurement. |
| def pyclustering.nnet.som.som.show_distance_matrix | ( | self | ) |
Shows gray visualization of U-matrix (distance matrix).
| def pyclustering.nnet.som.som.show_network | ( | self, | |
awards = False, |
|||
belongs = False, |
|||
coupling = True, |
|||
dataset = True, |
|||
marker_type = 'o' |
|||
| ) |
Shows neurons in the dimension of data.
| [in] | awards | (bool): If True - displays how many objects won each neuron. |
| [in] | belongs | (bool): If True - marks each won object by according index of neuron-winner (only when dataset is displayed too). |
| [in] | coupling | (bool): If True - displays connections between neurons (except case when function neighbor is used). |
| [in] | dataset | (bool): If True - displays inputs data set. |
| [in] | marker_type | (string): Defines marker that is used to denote neurons on the plot. |
| def pyclustering.nnet.som.som.show_winner_matrix | ( | self | ) |
Show a winner matrix where each element corresponds to neuron and value represents amount of won objects from input data-space at the last training iteration.
| def pyclustering.nnet.som.som.simulate | ( | self, | |
| input_pattern | |||
| ) |
Processes input pattern (no learining) and returns index of neuron-winner.
Using index of neuron winner catched object can be obtained using property capture_objects.
| [in] | input_pattern | (list): Input pattern. |
| def pyclustering.nnet.som.som.size | ( | self | ) |
| def pyclustering.nnet.som.som.train | ( | self, | |
| data, | |||
| epochs, | |||
autostop = False |
|||
| ) |
Trains self-organized feature map (SOM).
| [in] | data | (list): Input data - list of points where each point is represented by list of features, for example coordinates. |
| [in] | epochs | (uint): Number of epochs for training. |
| [in] | autostop | (bool): Automatic termination of learning process when adaptation is not occurred. |
| def pyclustering.nnet.som.som.weights | ( | self | ) |