 |
 |
Class implements G-Means clustering algorithm. More...
Public Member Functions | |
| def | __init__ (self, data, k_init=1, ccore=True, kwargs) |
| Initializes G-Means algorithm. More... | |
| def | process (self) |
| Performs cluster analysis in line with rules of G-Means algorithm. More... | |
| def | predict (self, points) |
| Calculates the closest cluster to each point. More... | |
| def | get_clusters (self) |
| Returns list of allocated clusters, each cluster contains indexes of objects in list of data. More... | |
| def | get_centers (self) |
| Returns list of centers of allocated clusters. More... | |
| def | get_total_wce (self) |
| Returns sum of metric errors that depends on metric that was used for clustering (by default SSE - Sum of Squared Errors). More... | |
| def | get_cluster_encoding (self) |
| Returns clustering result representation type that indicate how clusters are encoded. More... | |
Class implements G-Means clustering algorithm.
The G-means algorithm starts with a small number of centers, and grows the number of centers. Each iteration of the G-Means algorithm splits into two those centers whose data appear not to come from a Gaussian distribution. G-means repeatedly makes decisions based on a statistical test for the data assigned to each center.
Implementation based on the paper [17].
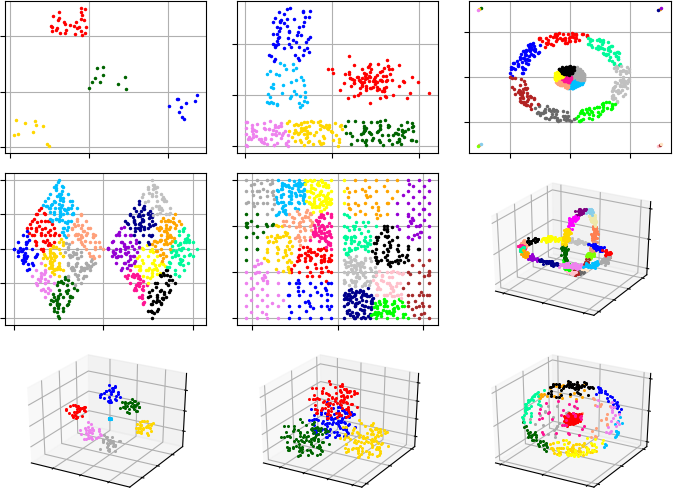
Example #1. In this example, G-Means starts analysis from single cluster.
Example #2. Sometimes G-Means might find local optimum. repeat value can be used to increase probability to find global optimum. Argument repeat defines how many times K-Means clustering with K-Means++ initialization should be run in order to find optimal clusters.
In case of requirement to have labels instead of default representation of clustering results CLUSTER_INDEX_LIST_SEPARATION:
There is an output of the code above:
| def pyclustering.cluster.gmeans.gmeans.__init__ | ( | self, | |
| data, | |||
k_init = 1, |
|||
ccore = True, |
|||
| kwargs | |||
| ) |
Initializes G-Means algorithm.
| [in] | data | (array_like): Input data that is presented as array of points (objects), each point should be represented by array_like data structure. |
| [in] | k_init | (uint): Initial amount of centers (by default started search from 1). |
| [in] | ccore | (bool): Defines whether CCORE library (C/C++ part of the library) should be used instead of Python code. |
| [in] | **kwargs | Arbitrary keyword arguments (available arguments: tolerance, repeat, k_max, random_state). |
Keyword Args:
k_max is -1).None, current system time is used). | def pyclustering.cluster.gmeans.gmeans.get_centers | ( | self | ) |
Returns list of centers of allocated clusters.
| def pyclustering.cluster.gmeans.gmeans.get_cluster_encoding | ( | self | ) |
Returns clustering result representation type that indicate how clusters are encoded.
| def pyclustering.cluster.gmeans.gmeans.get_clusters | ( | self | ) |
Returns list of allocated clusters, each cluster contains indexes of objects in list of data.
Definition at line 232 of file gmeans.py.
Referenced by pyclustering.samples.answer_reader.get_cluster_lengths(), and pyclustering.cluster.optics.optics.process().
| def pyclustering.cluster.gmeans.gmeans.get_total_wce | ( | self | ) |
Returns sum of metric errors that depends on metric that was used for clustering (by default SSE - Sum of Squared Errors).
Sum of metric errors is calculated using distance between point and its center:
![\[error=\sum_{i=0}^{N}distance(x_{i}-center(x_{i}))\]](../../form_9.png)
| def pyclustering.cluster.gmeans.gmeans.predict | ( | self, | |
| points | |||
| ) |
Calculates the closest cluster to each point.
| [in] | points | (array_like): Points for which closest clusters are calculated. |
| def pyclustering.cluster.gmeans.gmeans.process | ( | self | ) |
Performs cluster analysis in line with rules of G-Means algorithm.
Definition at line 165 of file gmeans.py.
Referenced by pyclustering.cluster.gmeans.gmeans.get_cluster_encoding().